Background
In a prvious post, I used GNfinder on a dozen or so sections of the Proceedings of the Academy of Natural Sciences of Philadelphia (ANSP) volumes that had been picked because they are likely to have a lot of taxonomic names in them. After I ID’d the genus and species names with GNfinder, I saved the list from each ANSP section as an entity set for use in spaCy, a natural language processing tool. Within spaCy (which we will run in a Jupyter Notebook b/c I still can’t make it work in R), I’ll run named entity recognition on the species names, and also look for locations and habitats.
In this exercise, I’ll run EVERY VOLUME of the ANSP corpus through GNfinder, and save the results in ONE GIANT entity list for spaCy. WEEE-HAW!! Let’s get wild.
Workspace setup
Load the Libraries we need.
Install and configure GNfinder
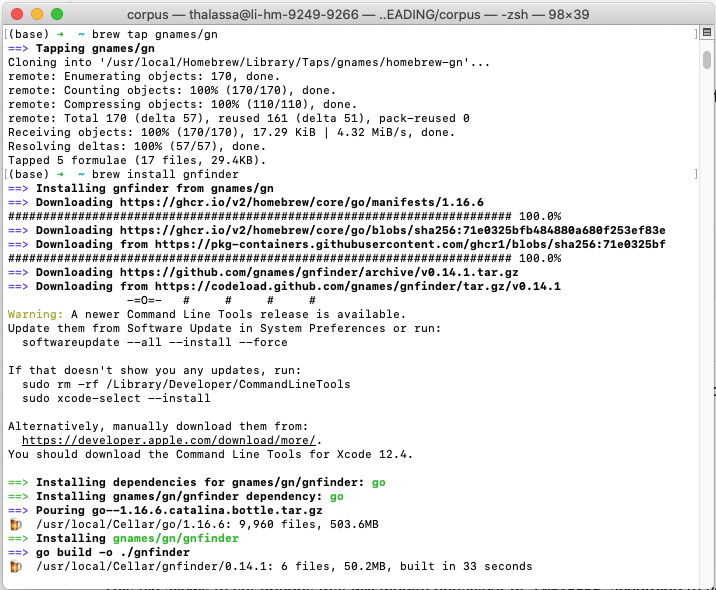
Recall that you need to have GNfinder installed on your machine to run it.
To run GNfinder1, see the instructions here. I am a Mac user, and I used the Homebrew option.
Create entity ruler for spaCy based on GNfinder results.
spaCy entity rulers look like:
{“label”: “HABITAT”, “pattern”: “temperate oceans”}
{“label”: “HABITAT”, “pattern”: “temperate reefs”}
{“label”: “HABITAT”, “pattern”: “temperate seas”} …
prefix <- str_c("{","\"label\": ","\"TAXA\","," \"pattern\": \"") # this looks ugly, but if we view it ...
# cat(prefix) # it's doing what we want.
sec1ents <- str_c(prefix,dat[[2]],"\"}") # put it together
# cat(sec1ents) # look at it - should be one entity (taxonomic name) per line, in the right format
Find taxonomic names in batch mode
We scraped the text (yay!), and now we want to feed that text, one file at a time, through the GNfinder tool. GNfinder will run in a Bash code chunk. I don’t know how to do this elegantly, so I’m just going to put all of the files I want to run into a folder and point GNfinder at it. If you have a more targeted way to make this work, say by using a list of filenames provided in a text file, please let me know. This feels a bit klugey to me.
Running GNfinder
Run gnfinder on a section of the Proceedings of the Academy of Natural Sciences of Philadelphia. Reminder: this runs in a bash chunk.
for f in /Users/thalassa/Rcode/blog/data/*.txt
do
echo "Processing $f file..."
gnfinder "$f" --verify > "${f%.txt}_gnf.txt"
doneCreate entity lists for use in spaCy NER pipeline
Now we need to read back in the results from GNfinder and use the taxonomic names it found to create an entity file for each one (in jsonl). Recall that we already created the ‘prefix’ to the pattern that we need in the entity file.
ldf <- list() # creates a list
listcsv <- dir(pattern = "*gnf.txt") # creates the list of all the csv files in the directory
outlist <- gsub(".txt", ".jsonl", listcsv ) #change the file extension for the entity files
for (k in 1:length(listcsv)){
dat <- as_tibble(read_csv(listcsv[[k]]))
entout <- str_c(prefix,dat[[2]],"\"}") # put it together
myfile <- str_c("/Users/thalassa/Rcode/blog/data/taxa-",outlist[[k]])
write_lines(entout, myfile)
}
rm(ldf, listcsv, outlist, dat, k, entout, myfile) # clean up!
Conclusion
Now we have the scraped text of 13 subsections of the ANSP corpus, we identified all of the txaxnomic names in each subsection using GNfinder, and we saved those taxa to text files (jsonl) that are formatted for use as spaCy entity sets in the NER pipeline. Now it’s time to return to a Jupyter Notebook to run these through! Stay tuned for that ….
Here are some screen shots that show you what the process resulted in.
Here are the files we created.
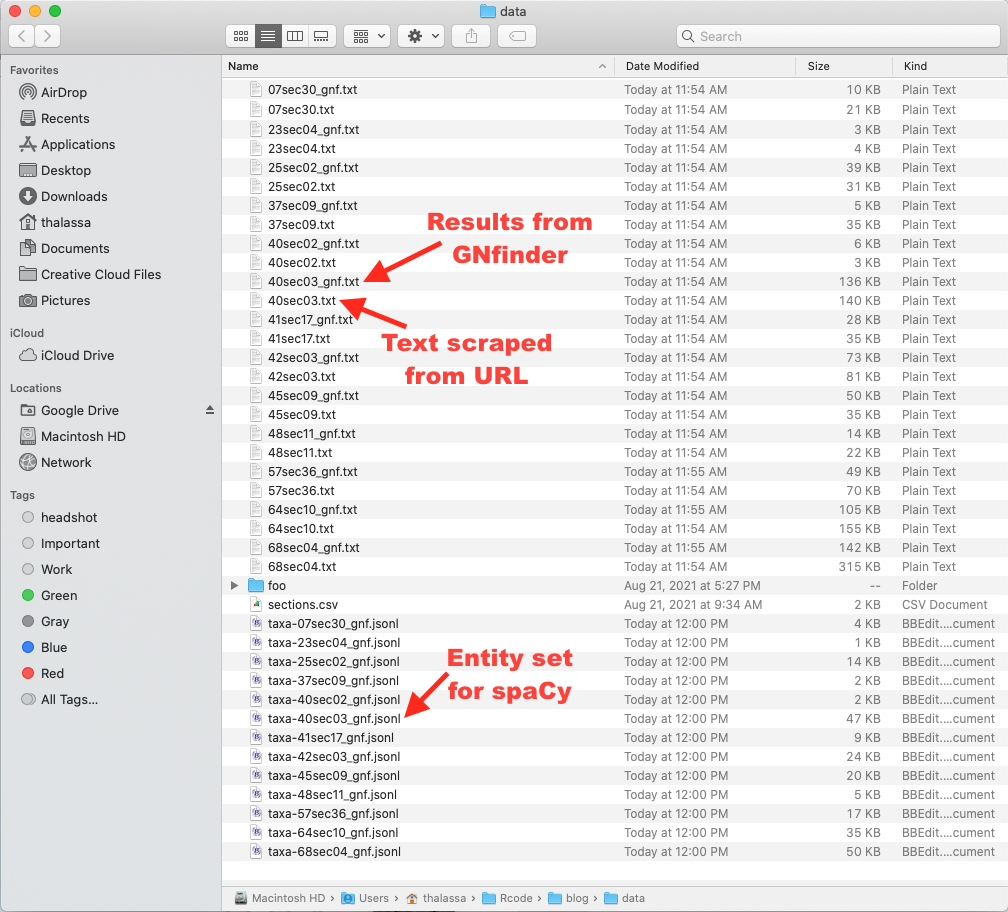
Here is what the ANSP OCR text looks like.
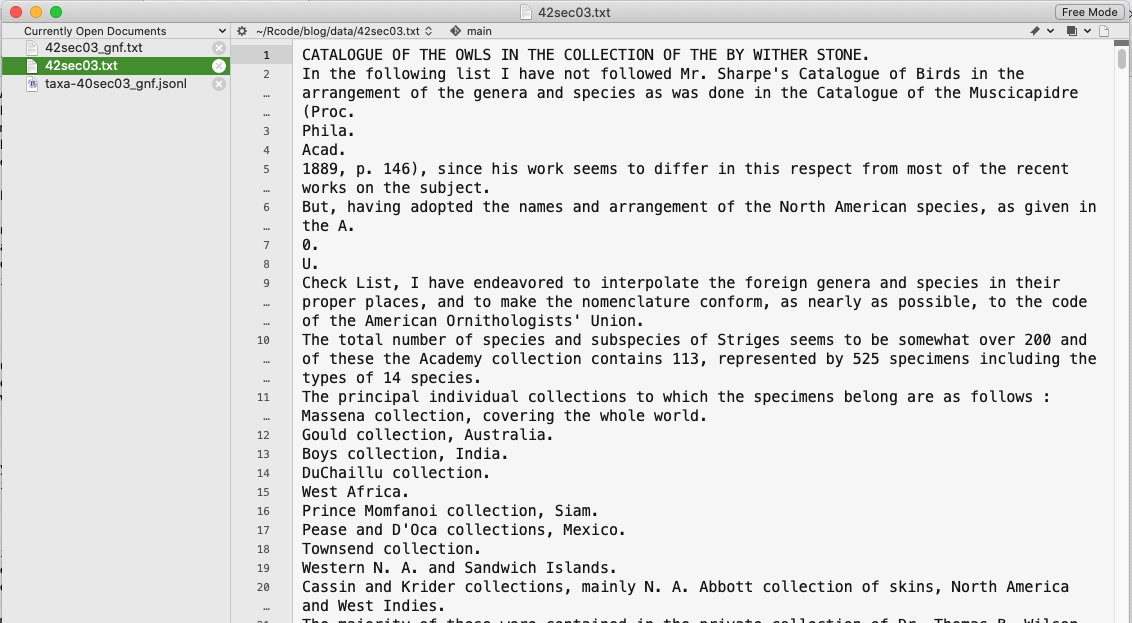
This is what the ouput of GNfinder looks like.
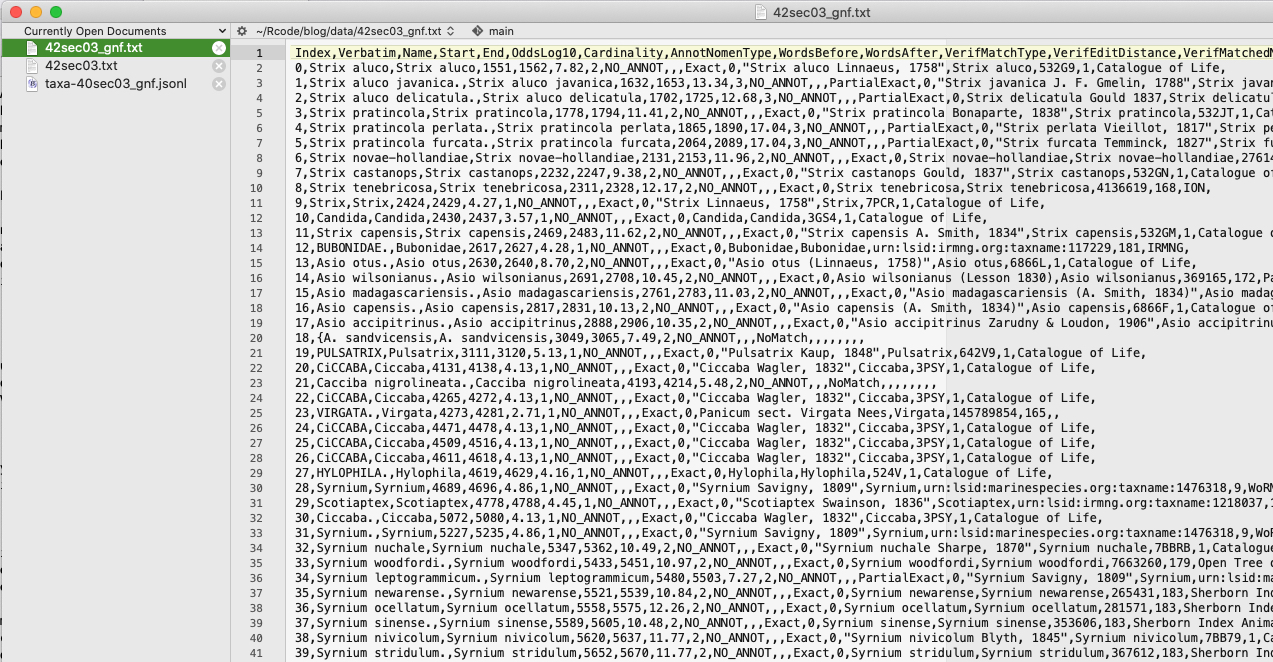
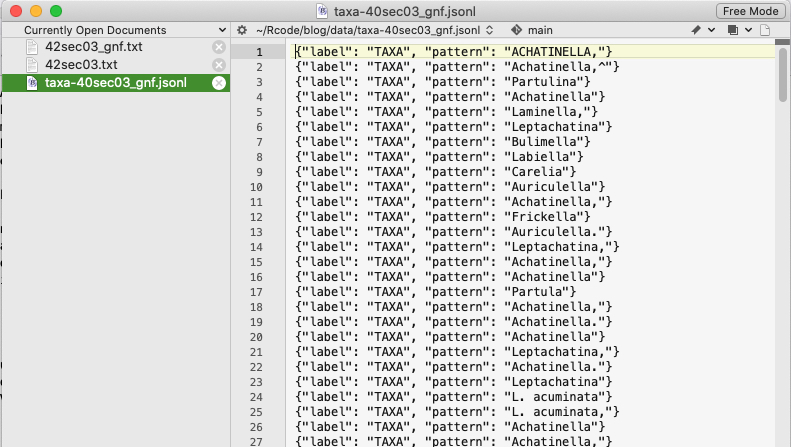
This is what the entity file for spaCy looks like.
Mozzherin, Dmitry, Alexander Myltsev, and Harsh Zalavadiya. Gnames/Gnfinder: V0.14.2. Zenodo, 2021. https://doi.org/10.5281/zenodo.5111562.↩︎