Background
In a previous post, I introduced myself to the Global Names Architecture tool Global Names Finder (GNfinder). In this post, I’m going to use GNfinder again, applying it on 134 volumes of the Proceedings of the Academy of Natural Sciences of Philadelphia (ANSP). After I identify the genus and species names, we’ll save the list from each ANSP section as an entity set for use in spaCy, a natural language processing tool. Within spaCy, we’ll run named entity recognition on the species names, and also look for locations and habitats. Onward!
Workspace setup
Load the Libraries we need.
Install and configure GNfinder
Recall that you need to have GNfinder installed on your machine to run it.
To run GNfinder1, see the instructions here. I am a Mac user, and I used the Homebrew option.
Find taxonomic names in batch mode
Real talk - we could do this by scraping text from BHL (see previous post), but I have all of the volumes as text files stored locally on my machine (DownThemAll Firefox browser extension - highly recommend), so I’m going to run this particular task locally (except it’s in Google Drive, but whatever). We want to feed that text, one file at a time, through the GNfinder tool. GNfinder will run in a Bash code chunk. I don’t know how to do this elegantly, so I’m just going to put all of the files I want to run into a folder and point GNfinder at it. If you have a more targeted way to make this work, say by using a list of filenames provided in a text file, please let me know.
Running GNfinder
Run gnfinder on a section of the Proceedings of the Academy of Natural Sciences of Philadelphia. Reminder: this runs in a bash chunk.
for f in /Users/thalassa/Google\ Drive/My\ Drive/LEADING/corpus/*.txt
do
echo "Processing $f file..."
gnfinder "$f" --verify > "${f%.txt}_gnf.txt"
doneCreate entity lists for use in spaCy NER pipeline
Now we need to read back in the results from GNfinder and pull out the verbatim taxonomic names it found to create lists for each volume. We’ll merge and de-duplicate these in following steps.
# creates the list of all the csv files in the directory
listcsv <- list.files("/Users/thalassa/Google Drive/My Drive/LEADING/corpus/gnfinder",
pattern = "*gnf.txt", full.names = T, recursive = TRUE)
outlist <- gsub("gnf.txt", "taxa.txt", listcsv) #change the file name for the entity files
for (k in 1:length(listcsv)){
dat <- as_tibble(read_csv(listcsv[[k]]))
myfile <- outlist[[k]]
write_lines(dat[[2]], myfile)
}
rm(listcsv, outlist, dat, k, myfile) # clean up!
That’s it - there is now a txt file with taxonomic names for every volume of the ANSP corpus. For the purposes of pulling identified taxonomic names into a generalized spaCy pipeline across any given section of the corpus, we’ll want one YUGE jsonl file with all of the results we just created. We need to merge, clean up gremlins and de-duplicate. Let’s do it!
#list all the files in the folder
listfile <- as_tibble(list.files("/Users/thalassa/Rcode/blog/data/taxa-verbatim-txt",
pattern = ".txt", full.names = T, recursive = FALSE))
data <- as_tibble(read_lines(listfile[[1]][1]))
for (k in 2:length(listfile)){
dat <- as_tibble(read_lines(listfile[[1]][k]))
data <- bind_rows(data, dat)
}
write_csv(data, "/Users/thalassa/Rcode/blog/data/ansp-taxa.txt")
rm(ldf, listfile, dat, k) # clean up!
I took this resulting list of verbatim taxonomic names from GNfinder and opened the file in BBEdit to do some minor clean-up. Like, could I have regex’d this in R? Yeah, painfully. But, BBEdit is really great! And fast! Here is an example of what the raw text looks like:

Notice all of the distracting punctuation. We don’t need any of that. I used simple grep commands to find punctuaiton at the beginning and ending of lines and did a “replace all” with "".
^[[:punct:]] finds punctuation at the start of the line. [[:punct:]]$ finds punctuation at the end of a line.
After that very minor cleaning, I ran Text > Process Duplicate Lines... because there is no reason to search for the same taxonomic name twice. Now we have:
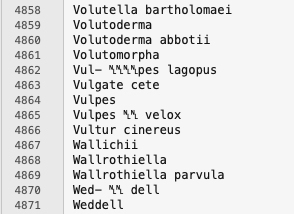
You’ll notice that there are still some odd characters inside of taxonomic names. Tempting to clean those up. VERY tempting. But, if we do that, we’ll actually miss finding taxonomic names as they appear in the text. We want our entities to reflect the corpus, not our hopes and dreams of what a cleaned corpus might look like. So, we leave it at that! Now I head back over into Jupyter Notebooks to get the NLP pipeline and a Streamlit app up and running …
Mozzherin, Dmitry, Alexander Myltsev, and Harsh Zalavadiya. Gnames/Gnfinder: V0.14.2. Zenodo, 2021. https://doi.org/10.5281/zenodo.5111562.↩︎